Hands-On Data Wrangling
Loading data
Following what we did last week, we are going to keep working with the penguins
today. You can find the dataset after you extracted session3.zip which
you can download from here.
Figure things out on the internet
If you don’t remember how to do something with R and happen to be the most R-fluent people in the peer, don’t panic. Most of the time, searching for [what you want to do] in R work out great.
Online forums like StackOverflow and Kaggle often give great answers with code examples that you could play with. If you are a genomic person, Biostar and SEQanswers would be your friend.
library(dplyr)##
## Attaching package: 'dplyr'## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union# How do you load a csv file with R?
penguins <- read.csv("penguins.csv")Figuring things out within R
Oftentimes, an answer you found online provides a code that almost works, but you might not yet know enough to make it work. For times like this, R has documentation built in for each function describing the arguments that you can tweak and what it meant.
You can trigger the help page with ?[function] in the console. Let’s try it
out: We used head() last week to take a glimpse of the first few rows of a
table, so how do we print the first 3 instead of 6 rows?
head(penguins, n = 3)## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18.0 195 3250
## sex year
## 1 male 2007
## 2 female 2007
## 3 female 2007count() things
How many penguins of each species were observed on each island?
penguins %>%
count(island, species)## island species n
## 1 Biscoe Adelie 44
## 2 Biscoe Gentoo 124
## 3 Dream Adelie 56
## 4 Dream Chinstrap 68
## 5 Torgersen Adelie 52Which species is found on all islands? I know it’s obvious, but let’s use a sledgehammer to crack a nut this time.
# The output in the previous chunk is also a table that could be count()ed.
penguins %>%
count(island, species) %>%
count(species)## species n
## 1 Adelie 3
## 2 Chinstrap 1
## 3 Gentoo 1filter() out your species
Let’s say you are interested in comparing how the islands influences the growth of penguins. Either you’ll need to go to Antarctica to observe more Gentoo or Chinstrap penguins on other islands, or only Adelie penguins make sense for your purpose.
# Since none of us is going to leave for Antarctica any time soon (right?)
# Let's keep only the Adelie penguins, and **assign it to a new object**.
adelie <- penguins %>%
filter(species == "Adelie")Revisiting the criteria for filter()
There are 6 basic types of comparison:
==: Equal to!=: Not equal to>: Larger than>=: Larger or equal to<: Less than<=: Less than or equal to
# How many male Adelie penguins were observed?
adelie %>%
filter(sex == "male") %>%
count()## n
## 1 73# How many Adelie penguins were found on islands that are not Dream island?
adelie %>%
filter(island != "Dream") %>%
count()## n
## 1 96# How many Adelie penguins were observed during or before 2008?
adelie %>%
filter(year <= 2008) %>%
count()## n
## 1 100Combining criteria
Sometimes, we’ll need more than one criteria to get the data we want. For example, if you are interested in the female Adelie penguins on the Biscoe island:
adelie %>%
filter(sex == "female", island == "Biscoe")## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## 1 Adelie Biscoe 37.8 18.3 174 3400
## 2 Adelie Biscoe 35.9 19.2 189 3800
## 3 Adelie Biscoe 35.3 18.9 187 3800
## 4 Adelie Biscoe 40.5 17.9 187 3200
## 5 Adelie Biscoe 37.9 18.6 172 3150
## 6 Adelie Biscoe 39.6 17.7 186 3500
## 7 Adelie Biscoe 35.0 17.9 190 3450
## 8 Adelie Biscoe 34.5 18.1 187 2900
## 9 Adelie Biscoe 39.0 17.5 186 3550
## 10 Adelie Biscoe 36.5 16.6 181 2850
## 11 Adelie Biscoe 35.7 16.9 185 3150
## 12 Adelie Biscoe 37.6 17.0 185 3600
## 13 Adelie Biscoe 36.4 17.1 184 2850
## 14 Adelie Biscoe 35.5 16.2 195 3350
## 15 Adelie Biscoe 35.0 17.9 192 3725
## 16 Adelie Biscoe 37.7 16.0 183 3075
## 17 Adelie Biscoe 37.9 18.6 193 2925
## 18 Adelie Biscoe 38.6 17.2 199 3750
## 19 Adelie Biscoe 38.1 17.0 181 3175
## 20 Adelie Biscoe 38.1 16.5 198 3825
## 21 Adelie Biscoe 39.7 17.7 193 3200
## 22 Adelie Biscoe 39.6 20.7 191 3900
## sex year
## 1 female 2007
## 2 female 2007
## 3 female 2007
## 4 female 2007
## 5 female 2007
## 6 female 2008
## 7 female 2008
## 8 female 2008
## 9 female 2008
## 10 female 2008
## 11 female 2008
## 12 female 2008
## 13 female 2008
## 14 female 2008
## 15 female 2009
## 16 female 2009
## 17 female 2009
## 18 female 2009
## 19 female 2009
## 20 female 2009
## 21 female 2009
## 22 female 2009What if you need penguins with extreme body weight? Say either over 4700g or below 2900g.
adelie %>%
filter(body_mass_g > 4700 | body_mass_g < 2900)## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## 1 Adelie Biscoe 36.5 16.6 181 2850
## 2 Adelie Biscoe 36.4 17.1 184 2850
## 3 Adelie Biscoe 41.0 20.0 203 4725
## 4 Adelie Biscoe 43.2 19.0 197 4775
## sex year
## 1 female 2008
## 2 female 2008
## 3 male 2009
## 4 male 2009Logical operations: What’s happening under the dplyr table
Essentially, what filter() relies on is to ask a series of yes/no question
to each row of a column, and the answers to these yes/no questions are called
Boolean or logical values.
In R, TRUE means yes, while FALSE means no.
# Is 3 larger than 5?
3 > 5## [1] FALSE# Is 2022 equal to 2020?
2022 == 2020## [1] FALSE# Is "apple" not equal to "orange"?
"apple" != "orange"## [1] TRUEThese logical values can be further calculated with & (AND), | (OR), and
xor().
& |
| |
xor() |
|
|---|---|---|---|
| TRUE/TRUE | TRUE | TRUE | FALSE |
| TRUE/FALSE | FALSE | TRUE | TRUE |
| FALSE/TRUE | FALSE | TRUE | TRUE |
| FALSE/FALSE | FALSE | FALSE | FALSE |
So, how many Adelie penguins either have a bill length shorter than 35mm or a bill depth below 15mm?
adelie %>%
filter(bill_length_mm < 35 | bill_depth_mm < 15) %>%
count()## n
## 1 9Now that we know filter() is working with logical values, you won’t be
surprised that it can also use other functions that gives a logical value
as its output as a criterion.
For example, we mentioned that NA is how R labels missing data. Since we are
interested in body weight, we might want throw away rows with missing body
weight…
# You might want to try filtering for body_mass_g== NA
adelie %>%
filter(body_mass_g == NA)## [1] species island bill_length_mm bill_depth_mm
## [5] flipper_length_mm body_mass_g sex year
## <0 rows> (or 0-length row.names)What is happening? Remember that we said R treats NA very differently. As a
matter of fact, almost every operation gives you NA when NA is involved.
# Try these
5 == NA## [1] NA3 > NA## [1] NA"North America" != NA## [1] NA3.1415926 <= NA## [1] NASince NA is missing data, this actually makes sense:
5 == NA is like asking “is 5 equal to something I don’t know?”, and the
answer has to be “I don’t know”.
So, how do we ask R if it doesn’t know something or has an NA there? We use
is.na().
is.na(NA)## [1] TRUEis.na("National Academy")## [1] FALSEYou can use is.na() with filter() to find the rows where body weight is
NA.
# Use is.na() to get rows with NA in body_mass_g
adelie %>%
filter(is.na(body_mass_g))## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## 1 Adelie Torgersen NA NA NA NA
## sex year
## 1 <NA> 2007How do we select all other rows then? In R, ! flips a logical value, and you
can pronounce it as not.
# Not TRUE
!TRUE## [1] FALSE# Not FALSE
!FALSE## [1] TRUE# Flipping (3 > 5)
!(3 > 5)## [1] TRUEWe can use ! to find rows that are not NA.
# How many Adelie penguins have their body mass recorded?
adelie %>%
filter(!is.na(body_mass_g)) %>%
count()## n
## 1 151Extended reading: Dealing with missing values
Depends on what you work with, you might not always want to drop missing values.
If you are not going to throw those data away, you’ll need to ask yourself questions like “Does missing value appear randomly?”, and decide how to best deal with them.
If you are interested, Hadley Wickham has a section in his book R for Data Science discussing this.
Group your data for further analysis
Let’s say we are interested in how body weight differs between islands. To perform this analysis, we might want to calculate the median and standard deviation of body mass per island.
To indicate how dplyr should group your data, we use group_by() to tell
it which column contains the group labels.
Let’s assign the grouped data to another object named adelie_per_island
adelie_per_island <- adelie %>%
filter(!is.na(body_mass_g)) %>%
group_by(island)Get summary statistics with summarize()
Now that we grouped the data, we can calculate the summary statistics.
The syntax for summarize() is as follows:
# You tell summarize() which functions to use on which column
adelie_per_island %>%
summarize(mean(body_mass_g))## # A tibble: 3 × 2
## island `mean(body_mass_g)`
## <chr> <dbl>
## 1 Biscoe 3710.
## 2 Dream 3688.
## 3 Torgersen 3706.By default, the column names of the summary table is the function call we gives
to summarize(), which could be a bit ugly. We can rename the columns by:
adelie_per_island %>%
summarize(mean_bw = mean(body_mass_g))## # A tibble: 3 × 2
## island mean_bw
## <chr> <dbl>
## 1 Biscoe 3710.
## 2 Dream 3688.
## 3 Torgersen 3706.The summary statistics that we use the most often are:
- Arithmetic mean (
mean()) - Median (
median()) - Standard deviation (
sd())
Let’s summarize the data with median and standard deviation per island.
# Summarize the Adelie subset per island with the median and standard deviation
# of the body mass
adelie_per_island %>%
summarize(bw_median = median(body_mass_g), bw_sd = sd(body_mass_g))## # A tibble: 3 × 3
## island bw_median bw_sd
## <chr> <dbl> <dbl>
## 1 Biscoe 3750 488.
## 2 Dream 3575 455.
## 3 Torgersen 3700 445.What if you don’t group?
adelie %>%
filter(!is.na(body_mass_g)) %>%
summarize(bw_median = median(body_mass_g), bw_sd = sd(body_mass_g))## bw_median bw_sd
## 1 3700 458.5661What if you want to group with another grouping variable?
# Let's group with sex
adelie %>%
filter(!is.na(body_mass_g), !is.na(sex)) %>%
group_by(sex) %>%
summarize(bw_median = median(body_mass_g), bw_sd = sd(body_mass_g))## # A tibble: 2 × 3
## sex bw_median bw_sd
## <chr> <int> <dbl>
## 1 female 3400 269.
## 2 male 4000 347.A picture is worth a thousand words
While it’s great to have summary statistics, oftentimes a quick visualization of our data will be very helpful.
qplot() provided by ggplot2 is designed for this purpose and can be easily
incorporated into your pipeline.
Example: a quick scatter/strip plot per island
Let’s plot something with points on a plane where the x-axis is the islands and the y-axis is the body weight.
# Load ggplot2
library(ggplot2)
adelie_per_island %>%
qplot(data = ., x = island, y = body_mass_g, geom = "point")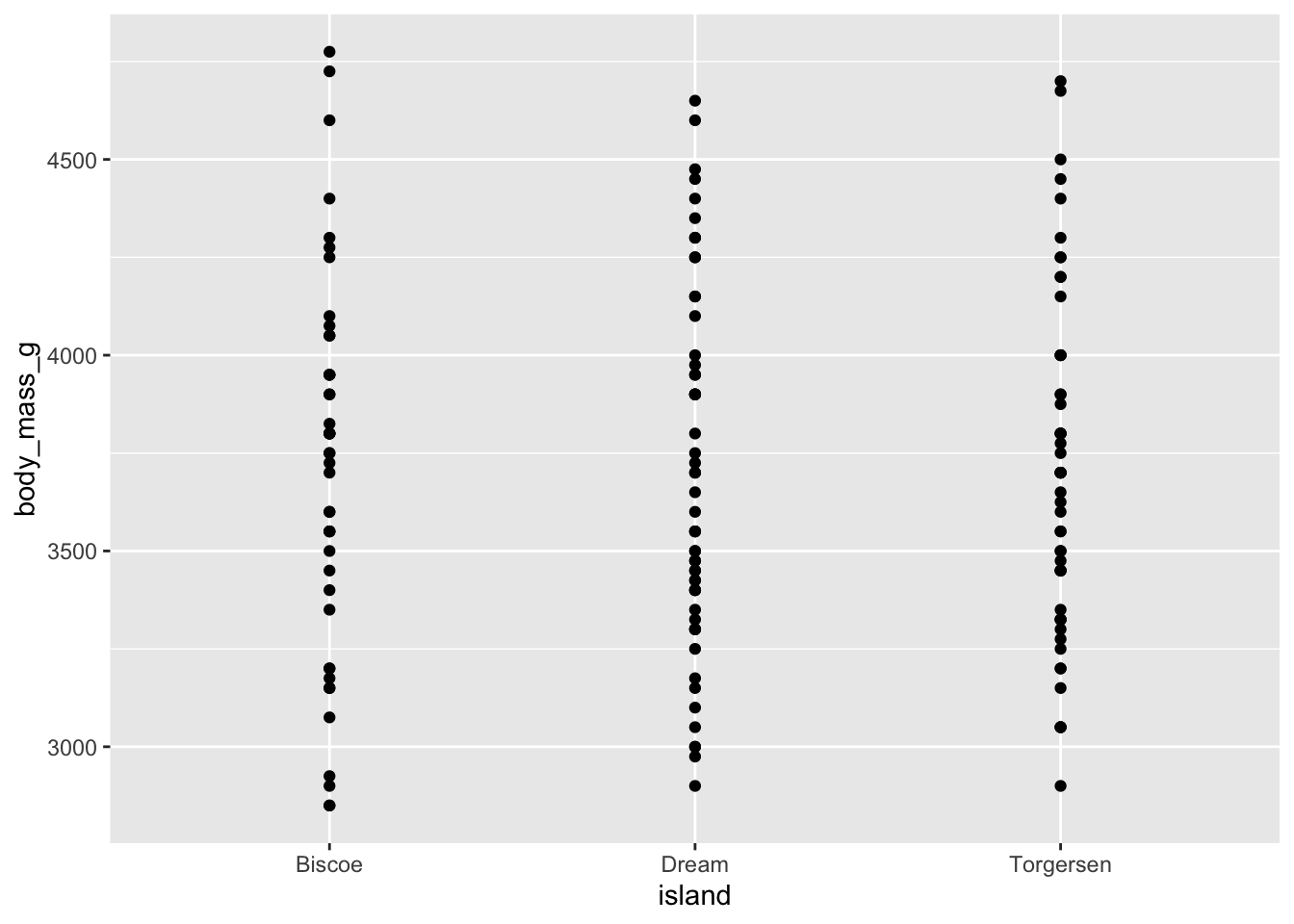
There several required arguments for qplot():
- data: The data you want to plot.
- x: The column name of the variable you want to use as the x-axis
- y: The column name of the variable you want to use as the y-axis
- color: The column name of the variable you want to color your plot by
- geom: Short for “geometry”, the type of plots you want to do. Popular ones
include:
- point
- bar
- boxplot
- violin
- histogram
- density
# Let's do a boxplot instead
adelie_per_island %>%
qplot(data = ., x = island, y = body_mass_g, geom = "boxplot")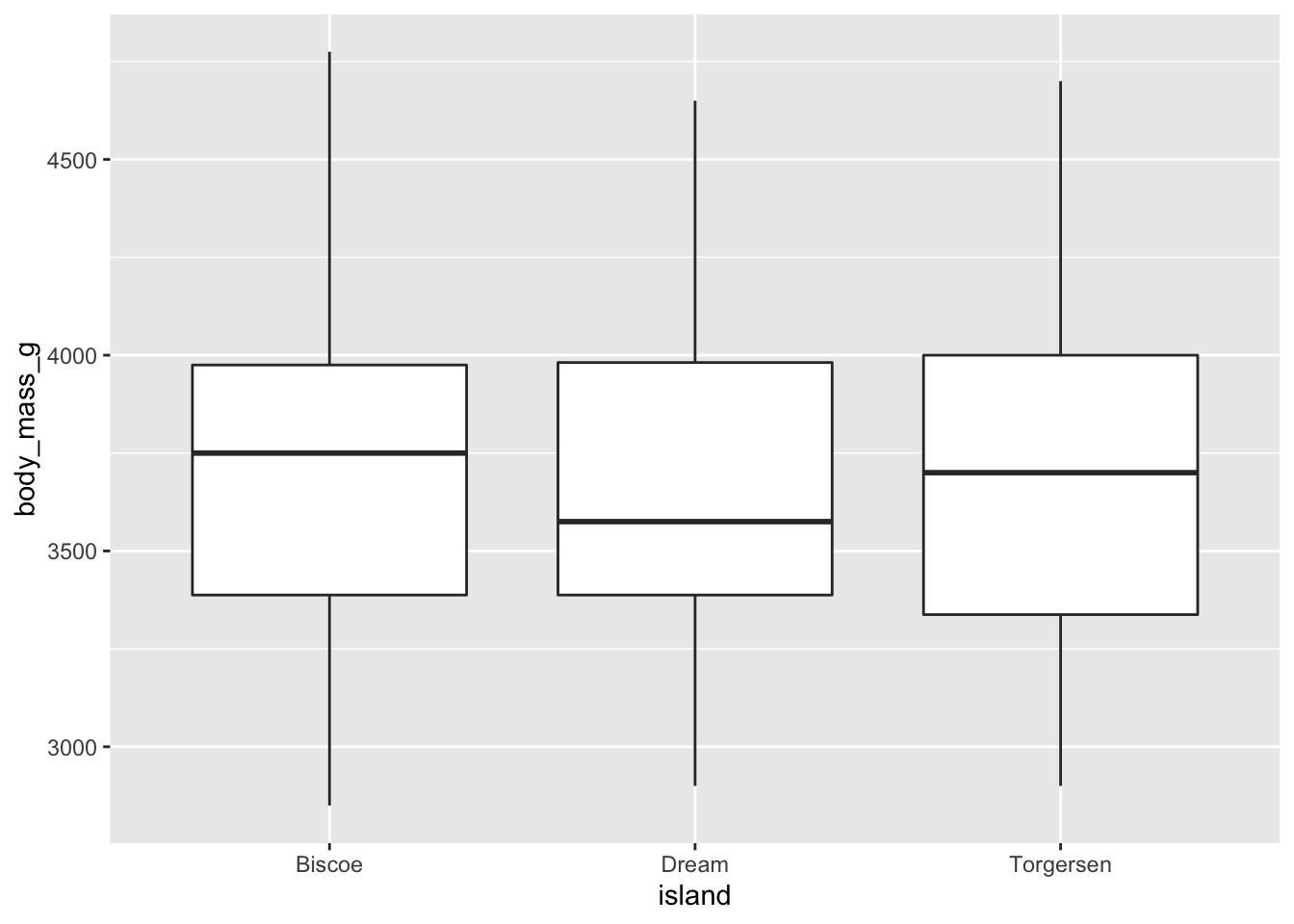 ### What is the distribution of bill lengths of the whole dataset?
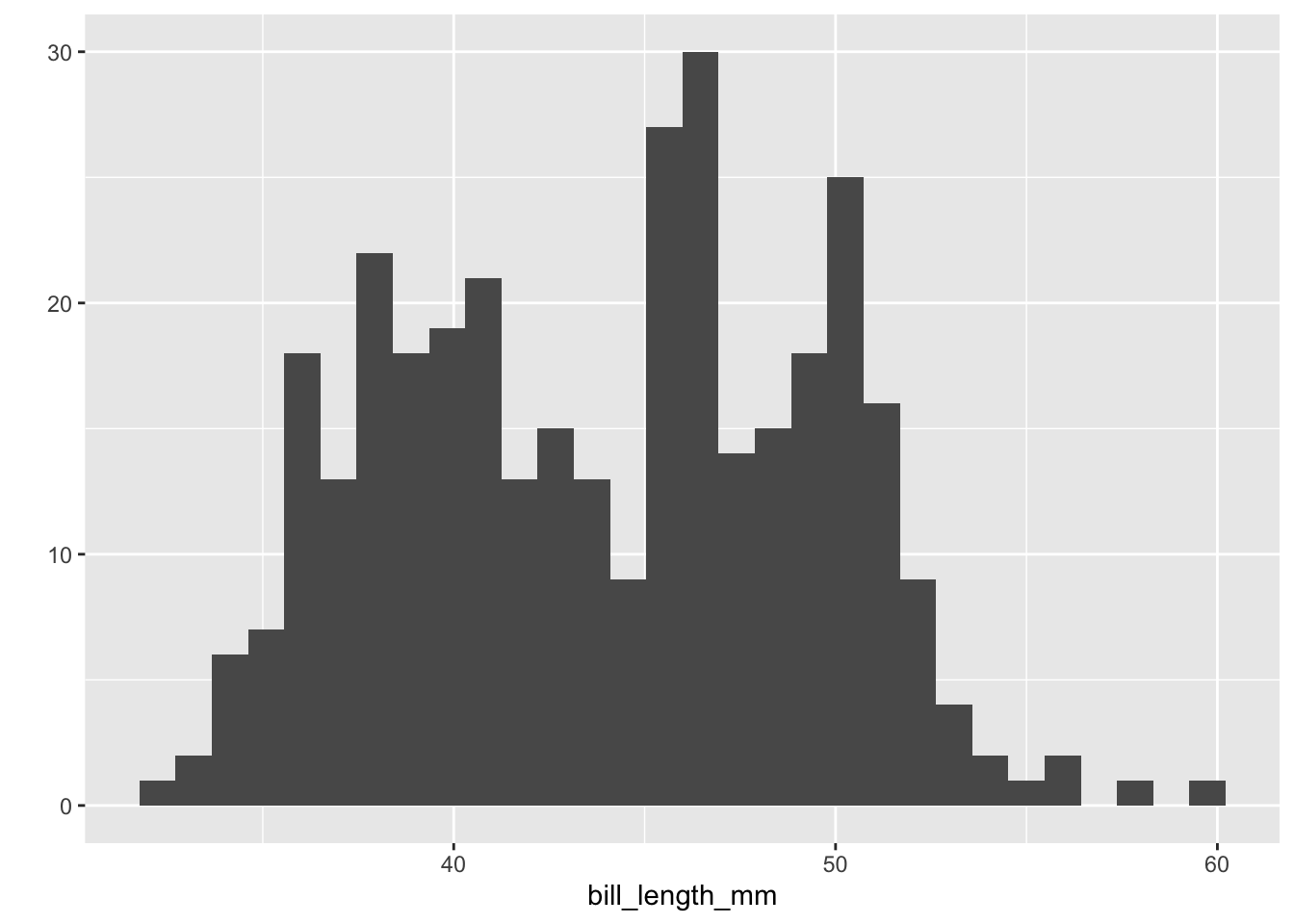
### What is the distribution of bill lengths of the whole dataset?
# Plot a histogram with bill lengths on the x-axis
penguins %>%
qplot(data = ., x = bill_length_mm, geom = "histogram")## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.## Warning: Removed 2 rows containing non-finite values (stat_bin).